Biophysics researcher investigates means to control the conformation of DNA molecules in nanopores
Sequencing the human genome allows for better understanding of genetic variations at the molecular level, such as the relationships among diseases, inheritance, and individuality. It increases the ability to take preemptive actions prior to disease development or to adopt treatments for diseases that have not yet been diagnosed.
In nanopore sequencing, single strands of DNA are driven by an applied electric field to pass through pores of nanometer size. Reducing a DNA molecule’s translocation speed in nanopores is a key step towards reliable resolution of individual bases.
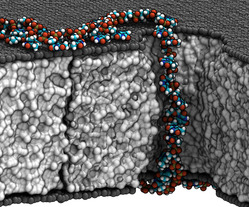
In a study aimed to explore means to control the conformation of DNA molecules passing through nanopores in a graphene membrane—atomic-scale hexagonal lattice made of carbon atoms—a group of University of Illinois researchers examined the effect of exposed graphene layers on the transport dynamics of both single- and double-stranded DNA through nanopores.
“Slowing DNA transport using graphene–DNA interactions” is a research project led by investigators from several departments across campus, including Center for Biophysics and Quantitative Biology Affiliate Professor Aleksei Aksimentiev (Department of Physics), Professor Rashid Bashir (Department of Electrical and Computer Engineering) and student researchers Shouvik Banerjee (Department of Materials Science and Engineering), James Wilson (Department of Physics), Jiwook Shim (Department of Electrical and Computer Engineering), Manish Shankla (Center for Biophysics and Computational Biology), and Elise A. Corbin (Department of Bioengineering).
While it was known that single-stranded DNA adsorbs hydrophobically to graphene, the research team hypothesized that hydrophobic adhesion to two such surfaces separated by a layer of inert material would stretch the molecule. “We’ve used all-atom molecular dynamics simulation to characterize the amount of stretching, thus showing that DNA nucleotides visit reproducible locations while passing through such double-graphene system,” says Professor Aksimentiev.
Nanopore devices with various combinations of graphene and dielectric layers in stacked membrane structures were fabricated as part of this study. The molecular dynamics simulations conducted confirmed the preferential interactions of DNA with the graphene layers as compared with the dielectric layer verifying the experimental findings.
Based on their findings, the researchers proposed that the integration of multiple stacked graphene layers could result in slowing down DNA sufficiently to place DNA nucleotides in reproducible configurations within the nanopore. According to Professor Aksimentiev, the stretching and controlled motion of DNA is essential to realizing solid-state nanopore sequencing, a technology that will further reduce the costs of DNA sequencing and may enable direct RNA sequencing, which is a major scientific and technological goal.
To read the full paper, please visit https://doi.org/10.1002/adfm.201403719.
Published July 22, 2019 20:01