Molecular mechanism of hearing highlighted in the first atomic-resolution picture of outer hair cell surface proteins
Our sense of hearing is stimulated by the sound transmitted through the external auditory canal to the middle ear and then to the inner ear. The hair cells in the inner ear are known as the sensory cells of hearing and are capable of mechano-electrical transduction—the mechanism by which cells convert a mechanical stimulus into an electrical signal—and signal amplification, which mechanically amplifies low-level sound entering the ear’s cochlea.
The outer hair cells (OHCs) of the cochlea play a major role in hearing sensitivity by providing the machinery for sound amplification. Without OHCs we would not be able to distinguish different sounds. At the heart of the electromechanical activity of OHCs, and their contraction and expansion in response to sound signals, is a motor protein, called prestin. Despite the discovery of prestin and its unique electromechanical functionality many years ago, and notwithstanding decades of extensive research, there has been no atomic resolution structures for the protein and little insight into its underlying mechanism.
University of Illinois at Urbana-Champaign researchers, in collaboration with researchers at the Oregon Health and Science University, led an investigation that was recently published in Cell to reveal, for the first time, the molecular mechanisms for cochlear amplification in mammalian hearing mediated by the electromotive activity of prestin. Capitalizing on the power of single-particle cryo-electron microscopy, advanced molecular dynamics simulation, and electrophysiology, the research team unveiled the structure and mechanism of prestin-mediated cochlear sound amplification.
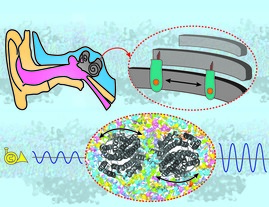
The study, led by Emad Tajkhorshid, Professor of Biochemistry and Center for Biophysics and Quantitative Biology affiliate, shows how prestin contraction and expansion in response to sound result in substantial changes in the cell, which in turn amplify the signal in OHCs.
“The study is a magnificent example of how combining different biophysical techniques—in this case, cryo-EM structural work and computational investigation of the system’s dynamics in an explicit membrane—could provide deep mechanistic insight that would be impossible to achieve otherwise,” says Professor Tajkhorshid.
“We are lucky at Illinois to have one of the best software programs, Nanoscale Molecular Dynamics (NAMD), for simulating the most complicated biological processes and the best visualization code, Visual Molecular Dynamics (VMD), to visually examine the system we study,” he adds.
Both programs have been developed at the NIH Center for Macromolecular Modeling and Bioinformatics at Beckman Institute for Advanced Science and Technology with long-term support from NIH (P41-104601).
Among the members of the research team are Biophysics and Quantitative Biology graduate students Sepehr Dehghani-Ghahnaviyeh and Zhiyu Zhao, both members of the Tajkhorshid lab.
According to Sepehr, employing the power of advanced computational techniques and cryo-EM enabled the investigators to shed light on something that had been puzzling biologist and biophysicists for decades. “I am so proud to be part of this work,” he says. Likewise, Zhiyu believes that this study demonstrates the power of computational methodologies and techniques in investigating the dynamics of biological systems, which, coherently with experimental efforts, can provide insights into their functions. “The publication arises from our close collaboration with Eric Gouaux's lab, and it's my pleasure to work with these great researchers,” he adds.
To review the complete findings of the study, you can access the published article on Cell webpage here.
Published September 10, 2021 16:17